Researchgroup of Professor Feng Chen, School of Public Health, NanjingMedical University
Lungcancer is the leading cause of cancer-related deaths in China and inthe world. It is well known that, in addition to environmentalexposures (e.g., tobacco smoking), single-nucleotide polymorphisms(SNPs) are one of the important genetic factor contributing to lungcancer susceptibility. Although many susceptible SNPs have beenidentified in genome-wide association studies (GWAS) during the pasttwo decades, together these identified SNPs still explain only alimited proportion of the heritability of lung cancer.
Lungcancer is a complex disease. The occurrence and development ofcomplex diseases are driven by complex association patterns, such asgene-environment(G×E)andgene-gene interaction(G×G).Statistically, the interaction of factors A and B indicates thatthere is synergy or antagonism between the two factors, or that theeffect of factor A changes with the value of factor B. Interaction isa phenomenon manifested at the population level through a complicatedpattern of association between two or more factors, providing anunusual way of thinking for in-depth understanding of the mechanismof disease occurrence and development. However, genome-wide gene-geneinteraction studies involve more than 50 billion tests. Thesuper-heavy computational burden greatly limits the feasibility ofgene-gene interaction research, therefore rational and efficientdimensionality reduction analysis strategies and methods forinteraction are urgently needed.
Recently,Professor Feng Chen from the School of Public Health of NanjingMedical University, academician Hongbing Shen, and Professor David C.Christiani from the Harvard School of Public Health in the UnitedStates have cooperated in depth and joined hands with theInternational Lung Cancer Research Alliance (ILCCO/TRICL) to completethe world's largest scale (465,000 people) study of gene-geneinteraction and trans-ethnic validation of lung cancer by adoptingthe three-step dimensionality reduction analysis strategy of'information entropy preliminary screening →log-linear modeltest →logistic regression model confirmation', and toconstruct an interaction-empowered polygenetic risk score (iPRS)simultaneously. Compared with the classic model of lung cancergenetic score (PRS-128) in the field, iPRS shows a better riskdiscrimination ability, which can further enhance the screeningability of high-risk groups of the internationally renowned lungcancer risk prediction model (LLPv3). The results were published asoriginal article in the Journal of Thoracic Oncology (IF=15.609,IMF=19.891, CAS/JCR Zone 1). Associate Professor Ruyang Zhang, Dr.Sipeng Shen and Associate Professor Yongyue Wei from the School ofPublic Health in Nanjing Medical University are the co-first authorsof the paper, Professor Feng Chen, Academician Hongbing Shen andProfessor David C. Christiani are the co-corresponding authors of thepaper.

Itadopts two designs in genome-wide G × G interaction study: atwo-phase design including discovery phase and validation phase,Meta-analysis (Figure1).(1)Bonferroni-corrected significant G × G interactions were identifiedin the discovery phase using ILCCO/TRICL from more than 59 billionpossible G ×G interactions considered in this study. Furthermore,these signals identified in the discovery phase were furtherconfirmed in the validation phase using UK Biobank as theverification set. (2) The genetic mechanism of lung cancer may alsobe different due to the heterogeneity of population with differentpathological tissue types, smoking status and gender. Therefore,researchers combined all GWAS samples and resorted to meta-analysisusing ILCCO-OncoArray, TRICL, and UK Biobank to identify on weakeffect G × G signals in the sub populations to identify lung cancersusceptibility loci.

Figure1. The workflow diagram of this study
Throughtwo-phase design, researchers identified two pairs of SNPs withantagonistic effects: rs2853668 of TERT gene at 5p15.33 andrs62329694 of CLPTM1L gene at 5p15.33, rs521828 of C6orf10 gene at6p21.32 and rs204999 of PRRT1 gene at 6p21.32. Furthermore, six SNPpairs with interaction signals were newly identified from specificsubpopulations through meta-analysis. Figure2summarizes the results of eight pairs of interaction signals.Bioinformatics analysis shows that these loci or genes may have richbiological functions.

Figure2. Summary of Genome-wide Gene-Gene Interaction Studies of LungCancer
Theresults of linkage disequilibrium (LD) analysis between loci indicatethat the five pairs of interactions in the 6p21.32 region are mainlydriven by two clusters of signals: rs521828-rs204999 andrs3135369-rs285859-rs9275572 (Figure3).

Figure3. Five pairs of interactions in the 6p21.32 region and LD
Basedon the ILCCO data, researchers further filled and analyzed 6p21.32and 5p15.33 regions. All SNPs within the approximately 500 KBflanking regions of the significant epistatic pairs were furthertested by logistic regression models to detect a cluster of G × Gsignals enriched in close proximity to the identified pairs (Figure4)

Figure4. The three-dimensional G × G interaction signal map in 6p21.32 and5p15.33 regions
Researchersfirstly evaluated the eight pairs of signals identified in thisinteraction study from the European-ancestry population by using anexternal Asian population. Conversely, only one pair of signals(2p32.2 region) among Asian population (NJMU-GSA) was validated byusing the European-ancestry population. Trans-ethnic validation showsthat the G × G interaction of lung cancer has both commonality andheterogeneity among different race (Figure5).

Figure5. The comparison of G × G interaction association results andeffect of allele frequency between Europeans and Asians
Furthermore,the researchers constructed iPRS in non-smokers and former smokersrespectively. Compared with PRS-128, which is well-known in thefield, iPRS has a better discrimination power of lung cancer risks(Figure6A-B).And the iPRS enhanced model can be served as a good risk classifierto identify high-risk groups of lung cancer effectively (Figure6C-D).This effect has been validated in independent populations.

Figure6. IPRS lung cancer risk discrimination ability and high-riskpopulation screening ability
Low-dosecomputed tomography (LDCT) has been proved to be an effective measurefor lung cancer screening. Currently U.S. Preventive Services TaskForce (USPSTF) and the Centers for Medicare and Medicaid Services(CMS) recommend LDCT screening to target subjects merely on the basisof their age and smoking pack-years. Clinically, the iPRS enhancedmodel may change the practice of lung cancer screening. For example,subjects aged less than 55 years or smoked less than 30 pack-years,but with a high iPRS, may be suggested as the high-risk populationfor lung cancer screening; for those with a high iPRS (>80%quartile) and smoked more than 60 pack-years, lung cancer screeningmay be better to start before 50 years old; and for those with a lowiPRS (<40% quartile), screening allowed to be postponed (Figure7).

Figure7. The absolute lung cancer incidence rates were presented forsubjects at different iPRS, pack-years, and age groups
Toimprove the cost-effectiveness of lung cancer screening, more than 20lung cancer risk prediction models have been created in the lastcouple of decades, by incorporating more clinical factors todistinguish high-risk populations. Among them, the Liverpool LungProject lung cancer risk model version 3 (LLPv3) is a well-known andvalidated model for lung cancer risk stratification. In addition, theresearchers found that iPRS could further stratify subjects intodifferent risk groups even at the same subgroup stratified by LLPv3,implying iPRS could enhance the screening ability to identifyhigh-risk groups of lung cancer (Figure8).
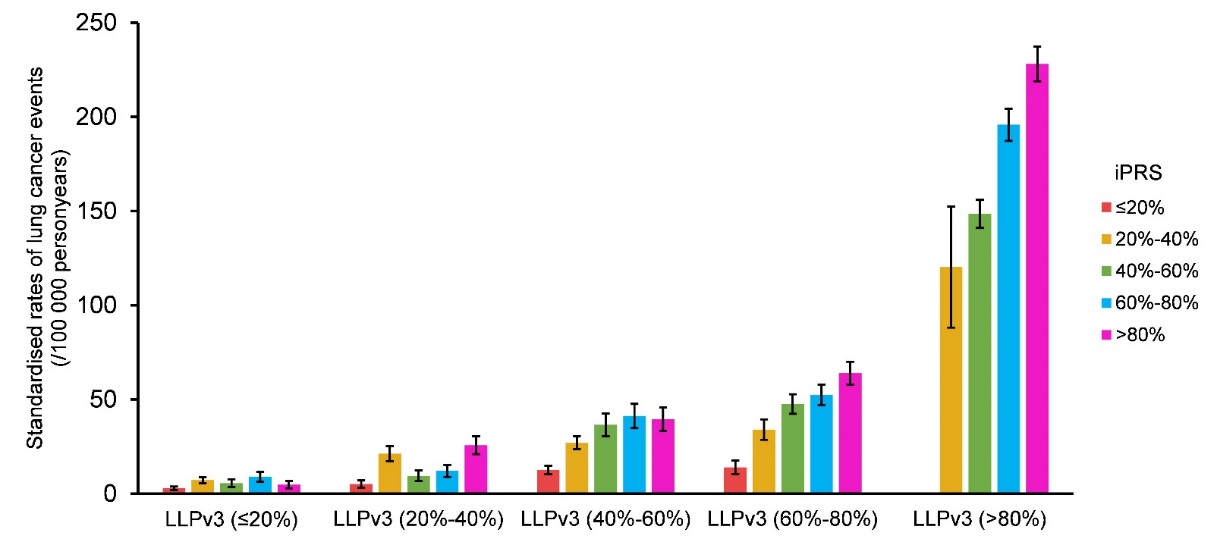
Figure8. Incidence rate of lung cancer in different LLPv3 and iPRS groups
ProfessorFeng Chen and Academician Hongbing Shen's research team have along-term focus on the interaction study of lung cancer, andpublished the first report of genome-wide gene-gene (Chu M, et al.Carcinogenesis. 2014) and gene-smoking (Zhang R, et al.Carcinogenesis. 2014) interaction study of lung cancer in Asianpopulation as a co-corresponding author. Professor Christopher I.Amos is the director of the Clinical Transformation ResearchInstitute of Baylor College of Medicine, one of the founders ofILCCO/TRICL, two top global lung cancer research alliances, and alsoa leader in international lung cancer genetic statistics andepidemiology. In his highly cited Web of Science paper, A decade ofGWAS results in lung cancer, he respectively evaluated the researchon gene-gene and gene-smoking interaction of lung cancer in Asianpopulations as 'the only research on gene-gene interaction atthe omics level so far' and 'the first research ongene-smoking interaction at the omics level' (Boss é Y, et al.Cancer Epidemiology, Biomarkers & Prevention. 2018).
Thisstudy which is of great significance is not only the largest G × Ginteraction study of lung cancer risk in the world, but also thefirst G × G interaction GWAS among the European-ancestry populationwith trans-ethnic validation.
Greatthanks to the International Lung Cancer Research Alliance(ILCCO/TRICL) led by Professor Christopher I. Amos of Baylor Collegeof Medicine and Professor Rayjean J. Hung of the University ofToronto, Canada, and all alliance members for their full support.
Time:May, 2022
Journalof Thoracic Oncology
Title:A large-scale genome-wide gene-gene interaction study of lung cancersusceptibility in Europeans with a trans-ethnic validation in Asians
Link:https://www.jto.org/article/S1556-0864(22)00215-5/fulltext

